1. Introduction
Doenjang (soybean paste) is a unique traditional fermented food from South Korea, which is produced by the natural growth of microorganisms, such as bacteria, yeasts, and fungi, in meju (Korean fermented soybean koji), followed by fermentation and ripening (Seon et al., 2021). The most dominant fungal species in doenjang is Aspergillus oryzae, which is a Generally Recognized as Safe (GRAS) strain and a microbial species that has been extensively consumed by humans. This species has been used as the main fermentation starter in the production of meju and nuruk because of the strong degradation activities of enzymes such as protease and amylase (Eaton and Gallagher, 1994; Lee et al., 2014; Park et al., 2001). However, the fungal species A. flavus, which exhibits high morphological and genetic similarities to A. oryzae, is frequently found in traditional Korean fermented foods, such as meju and doenjang etc., and the fungal toxins produced by these species have become a social issue (Jung et al., 2012; Kwon et al., 2011a; Shukla et al., 2014).
A. flavus produces aflatoxin, which is a toxic secondary metabolite that can cause cancer and genetic mutations (Alshannaq and Yu, 2020; Payne et al., 2006). The four most common aflatoxins isolated from not only fermented foods and cereal grains but also animal feeds, are aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1), and aflatoxin G2 (AFG2). These toxins are classified as Group I carcinogens, which are carcinogenic to humans, by the International Agency for Research on Cancer (IARC) (Loomis et al., 2018; Park et al., 2002; Richard, 2007). Among these toxins, AFB1 has a higher detection frequency than the others in foods, such as cereal grains and feeds, accounting for 75% of aflatoxin contamination (Shivachandra et al., 2003). Moreover, it is considered as carcinogenic and responsible for the highest mutation rate (Eaton and Gallagher, 1994). Therefore, the World Health Organization (WHO), United Nations Food and Agriculture Organization (UN FAO), European Food Safety Association (EFSA), and several other public health care authorities in South Korea and internationally, have implemented strict criteria for fungal toxins that can cause various diseases and recommend their management and continuous monitoring (Kang et al., 2010; Moretti et al., 2017; Park et al., 2008).
A. oryzae and A. flavus are part of the same Aspergillus section Flavi and share highly similar characteristics (Chang and Ehrlich, 2010). These species have conventionally been distinguished by morphological and cultural characteristics rather than biochemical or genetic characteristics (Jørgensen, 2007). However, because several researchers have demonstrated the challenges of morphological differentiation of the Aspergillus section Flavi (Kjærbølling et al., 2020), the two species have been differentiated by gene sequencing in recent studies. Nevertheless, it has been reported that 10 in 200 isolated A. flavus strains had ⟩99% similarity with both A. flavus and A. oryzae. The remarkably similar phylogenetic relationship of the two species prevents complete differentiation based solely on phylogenetic analysis by partial sequencing, and therefore they may be used with a degree of uncertainty in terms of purity (Nargesi et al., 2021). Thus, a safety assessment regarding the potential production of fungal toxins by Aspergillus sp. is required because it is highly probable that fungal strains isolated from fermented foods with a high degree of similarity to A. oryzae will be used in food production without further safety evaluation owing to its incorrect identification as A. oryzae by simple microbiological identification methods (Lee et al., 2014).
Various methods are available for detecting fungal toxins such as aflatoxin, including thin-layer chromatography (TLC), enzyme-linked immunosorbent assay (ELISA), fluorescence detection (FLD), ultraviolet light diode array detector (UV/DAD), and high-performance liquid chromatography (HPLC) equipped with mass spectrometry (MS) (Hwang et al., 2004). Recently, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) has been extensively used as a method for simultaneous multi-component analysis. However, although LC-MS/MS is recommended by MFDS, not many laboratories are equipped with the instrument because of its high cost, complicated operation, and need for skilled technicians. Thus, quantification methods using common HPLC coupled with FLD, as recommended by the Association of Official Analytical Chemists (AOAC), or the rapid and facile ELISA kit are extensively used in research laboratories (Kim and Kim, 2012; Meneely et al., 2011). Therefore, this study aims to screen fungal strains of the Aspergillus genus regarding their safe use in fermented food production by comparing the performance of different analytical methods, including UPLC, HPLC, and ELISA, and evaluating the corresponding aflatoxin production.
2. Materials and methods
This study was conducted to compare the accuracy of quantification analyses for aflatoxin detection because of the difficulty in differentiating the fungal species isolated from doenjang, such as A. oryzae, and A. flavus, based solely on their morphological and genetic characteristics.
The six test strains included 4 out of A. oryzae strains (P3, P3, P5, and P7) isolated from doenjang and 2 out of A. oryzae strains (40-2 and 83-3) isolated from nuruk (Fig. 1). The aflatoxin producing positive strains were the follows; A. flavus strains (KACC46453, KACC46817, and KACC46449) isolated from meju and obtained from the Korean Agricultural Culture Collection (KACC). The aflatoxin non-producing negative strains were A. oryzae strains (ATCC1011 and RIB40) purchased from the American Type Culture Collection (ATCC) and the National Research Institute of Brewing (NRIB). All strains were stored in a 25% glycerol stock (w/v) at −80°C, and then applied to potato dextrose agar (PDA; BD, Franklin Lakes, NJ, USA) to culture for 3 days at 35°C for subsequent use.
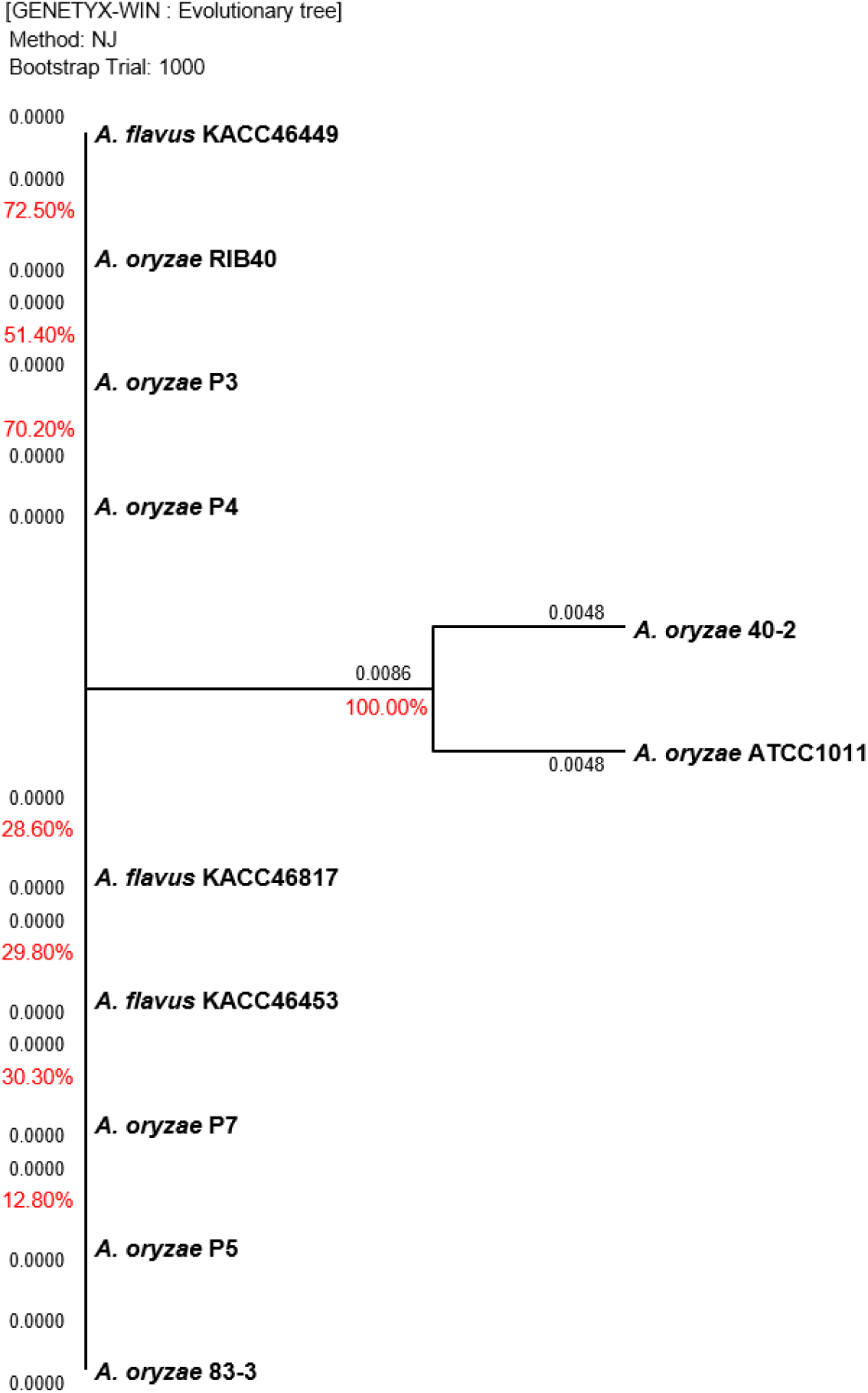
The Aspergillus sp. strains cultured on PDA at 35°C for 3 days were used for gene sequencing. Polymerase chain reaction (PCR) and sequencing were performed according to the manual (Macrogen Inc., Daejeon, Korea) using primers ITS5 (TCCG TAGGTGAACCTGCGG) and ITS4 (TCCTCCGCTTATT GATATGC) of the internal transcribed spacer (ITS) regions and primers NS1 (GTAGTCATATGCTTGTCTC) and NS24 (AAACCTTGTTACGACTTTTA) of the 18S ribosomal ribonucleic acid (18S rRNA) regions. Based on the sequencing data of Aspergillus sp., a phylogenetic tree was produced to generate clusters according to the genetic distance using the GENETYX-WIN (version 5) software with 1,000 bootstrapping trials.
For the pretreatment of aflatoxin for UPLC and HPLC analyses, the extraction and purification method of Lee et al. (2021) was modified and used. For extraction, 20 mL of extraction solution (70% methanol containing 1% NaCl) was added to 5 g of PDA with cultured Aspergillus sp., and the mixture was shaken at 300 rpm for 1 h. The resulting extract was centrifuged at 2,480 ×g for 10 min, and then 10 mL of supernatant was diluted with 30 mL of 1% Tween 20 (Sigma, St. Louis, MO, USA). Subsequently, 20 mL of the diluted solution was transferred to a purification column (AflaTestWB, Waters, Milford, MA, USA). To remove any impurities in the column, 10 mL of distilled water was used, after which aflatoxin was eluted by applying 2 mL of methanol (MeOH; HPLC grade, Fisher, Darmstadt, Germany). The eluate was dried in N2 at 40°C, and then mixed with 0.2 mL of trifluoroacetic acid (TFA; Sigma, St. Louis, MO, USA) for 15 min of reaction in the dark. Subsequently, 0.8 mL of 20% acetonitrile (ACN; HPLC grade, Fisher, Darmstadt, Germany) was added to the reaction mixture for dissolution, followed by filtration using a 0.2 μm syringe filter (PALL, Port Washington, NY, USA). Subsequently, UPLC and HPLC analyses were performed.
Quantitative analyses were performed on aflatoxin B1, B2, G1, and G2 by UPLC and HPLC. For UPLC (WATERS ACQUITY UPLC H Class, Waters, Milford, MA, USA) analysis, an Xselect CSH C18 column (2.5 μm, 2.1 mm, I.D. = 00 mm, Waters, Milford, MA, USA) was used under the following conditions: flow rate = 0.2 mL/min, run time = 10 min, column temperature = 40°C, injection volume = 10 μL, fluorescence detector (FL) wavelengths of Ex = 360 nm and Em = 440 nm, and mobile phase of ACN:MeOH:water = 15:20:65 (v/v/v). For HPLC (HITACHI Chromaster CM5000 Series, HITACHI, Tokyo, Japan) analysis, a LaChrom C18-AQ column (3 μm, 4.6 mm I.D. = 150 mm, HITACHI, Tokyo, Japan) was used under the following conditions: flow rate = 1.0 mL/min, run time = 20 min, column temperature = 40°C, injection volume = 10 μL, FL detector wavelengths of Ex = 365 nm and Em = 450 nm, and mobile phase of ACN: MeOH:water = 10:30:60 (v/v/v). Table 1 summarizes the detailed analytical conditions.
The total amount of aflatoxins (B1, B2, G1, and G2) was quantitatively analyzed using an ELISA test kit (AgraQuant Total Aflatoxin, Romer Labs, Getzersdorf, Austria). For extraction, 25 mL of 70% MeOH was added to 5 g of PDA with cultured Aspergillus sp., and the mixture was shaken at 300 rpm for 3 min. The extract was centrifuged at 6,523 ×g for 10 min, the supernatant was filtered using Whatman filter paper (No.1, Whatman, Maidstone, UK), and the filtrate was used as the final sample extract. Subsequently, 200 μL of conjugate solution and 100 μL of the reference or sample extract solution at each set concentration were added to each dilution well and mixed. After transferring 100 μL of each mixture to an antibody-coated well, the plate was cultured for 15 min at ambient temperature. Each compartment was washed five times with distilled water and dried by gentle tapping on a Wypall towel (Yuhan-kimberly, Seoul, Korea). After adding 100 μL of substrate solution to each antibody-coated well, the plate was cultured for 5 min at ambient temperature. Subsequently, 100 μL of stop solution was added to the culture solution, and an ultraviolet (UV) spectrophotometer (Synergy Mx, BioTek, Winooski, VT, USA) was used to estimate the level of aflatoxin at a wavelength of 450 nm.
For the method validation in this study, the limit of detection (LOD), limit of quantification (LOQ), linearity, repeatability, accuracy, and recovery were determined based on the CODEX Guideline (1995) and the Guideline of Standard Procedures of Testing Methods on Foods etc. (2016) of the National Institute of Food and Drug Safety Evaluation (NIFDS) at the Ministry of Food and Drug Safety (MFDS) (CODEX, 1995; MFDS, 2016). To test the linearity for aflatoxin, the reference material aflatoxin mix (Romer Labs, Getzersdorf, Austria) was diluted using ACN to prepare the reference solution. The calibration curve for the reference solution at each set concentration was produced according to the corresponding peak areas obtained by UPLC (0.025-10 μg/L) and HPLC (10-1,000 μg/L) analyses. For the ELISA assay, the calibration curve for the reference solution (0-20 μg/L) included in the test kit was produced. The correlation coefficient was obtained for the calibration curves of the three methods.
To validate the accuracy and precision of the analytical methods, different concentrations of aflatoxin were added to PDA without aflatoxin contamination and the tests were performed after pretreatment. For UPLC and HPLC analyses, aflatoxin B1 and G1 were added at concentrations of 125, 250, and 500 μg/kg and aflatoxin B2 and G2 were added at concentrations of 31.25, 62.5, and 125 μg/kg. For ELISA analysis, aflatoxin B1, B2, G1, and G2 were added at concentrations of 2, 5, and 10 μg/kg to perform the validation test for recovery. All tests were performed in triplicate, and the mean and relative standard deviation (RSD) were estimated to validate the recovery. The calibration curve slope (S) and SD (σ) were used to calculate the LOD and LOQ values as follows: LOD = 3.3 × σ/S and LOQ = 10 × σ/S.
The SPSS software (IBM SPSS Statistics 20, SPSS Inc., NY, USA) was used to perform the statistical analysis on the experimental data using analysis of variance (ANOVA) and the Sheffe’s post-hoc test. To evaluate the significance of the mean values, the level of significance was set at p⟨0.05.
3. Results and discussion
Gene sequencing was performed to compare the genetic distance between the 18s rRNA 1,771 bp and ITS 672 bp regions of Aspergillus sp., and the results are shown in Fig. 2 and Fig. 3, respectively. The sequencing data for the two regions did not clearly differentiate between A. oryzae and A. flavus. For the 18S rRNA regions, the highest similarity of 100% was observed between the 40-2 and ATCC1011 strains isolated from nuruk. For the ITS regions, a high similarity of 91.7% was observed between the P3 and P5 strains isolated from doenjang. Most fungal strains are identified by sequencing the two well-known molecular markers, ITS and 18s rRNA (Back, 2014; Cheong et al., 2013; Kim, 2011; Kim et al., 2012; Kwon et al., 2011b). However, the representative Aspergillus strains, including A. oryzae RIB40, A. flavus ATCC42149, and A. flavus NRRL3357, exhibited ∼99.5% genome homology and ∼98% protein homology in previous studies, which agrees with the results obtained in this study (Rank et al., 2012; Rokas et al., 2007). Furthermore, according to Nargesi et al. (2021), it is practically impossible to distinguish between two species that possess high sequence homology, such as A. oryzae and A. flavus, based solely on fungal barcode genes, namely ITS or β-tubulin (benA). Therefore, several researchers have investigated methods to distinguish between the two species by targeting species-specific genes, such as aflT, norA, verA, and cyp51A, and exploring other genes (Choi et al., 2021; Nargesi et al., 2021). However, the differentiation of the two species using molecular genetic methods such as whole genome sequencing remains challenging and requires high cost and time. Thus, it is hypothesized that the physiological characteristics of the two species should be analyzed based on more accurate and rapid in vitro analyses of aflatoxin production.
Table 2 lists the comparison of the recovery, LOD, and LOQ results for the three analytical methods, UPLC, HPLC, and ELISA. In this study, linearity was tested by estimating the correlation coefficient (R2) for the calibration curves within each concentration range, which were 0.9994-0.9999 for UPLC, 0.9642-1.000 for HPLC, and 0.9996 for ELISA (data not shown). The UPLC and ELISA results satisfy the MFDS criterion of ≥0.99, whereas the average result obtained for HPLC (0.97) satisfies the CODEX criterion of ≥0.95, but not the MFDS criterion. In the validation of recovery, all three methods exhibited high recovery of ≥70% for UPLC and HPLC and ≥75% for ELISA. For UPLC and HPLC, the recovery was higher (80%) for the tests performed using high rather than low concentrations. For ELISA, the recovery was high (≥85%) at a low concentration of 2 μg/kg, indicating that aflatoxin detection is possible at low concentrations in samples. Regarding recovery, UPLC and HPLC satisfy the CODEX criterion of 70-110% at ≥100 μg/kg, whereas ELISA satisfies both the CODEX and MFDS criteria of 70-100% at 1-10 μg/kg. In the validation of precision, the RSD ranged from a minimum of 0.12% to a maximum of 12.14%, satisfying the MFDS and CODEX guidelines (Alshannaq and Yu, 2020; Trucksess et al., 2008). The LOD and LOQ values were 0.055-0.159 and 0.168-0.483 μg/kg for UPLC, and 0.20-2.79 and 0.60-2.57 μg/kg for HPLC, respectively. This indicates that UPLC can detect and quantify aflatoxin at lower concentrations than HPLC.
The validation parameters for the reliability of the selected aflatoxin detection methods include LOD, LOQ, linearity, repeatability, reproducibility, and recovery (MFDS, 2016). Consequently, regardless of the amount of time required for extraction and analysis, UPLC and HPLC are highly effective methods in current research for analyzing both low and high concentrations of aflatoxin. In contrast, although ELISA is an easy-to-use enzyme-based immunoassay that allows rapid detection, it cannot accurately analyze aflatoxin from low to high concentrations, which reflect the wide concentration range in target foods and microbial samples (Kim and Kim, 2012). However, this study demonstrated that UPLC and HPLC are suitable for accurate quantitative analysis of aflatoxin from low (≤10 μg/kg) to high (≥100 μg/kg) concentrations, whereas ELISA is suitable for more rapid quantitative analysis of aflatoxin at low concentrations (≤20 μg/kg).
Table 3 lists the quantification of aflatoxin production by Aspergillus sp. using the three analytical methods, UPLC, HPLC, and ELISA. Among the investigated strains (P3, P4, P5, and P7) isolated from doenjang, P5 and P7 exhibited high aflatoxin production, which exceeded the CODEX criterion (≤15 ppb in meju, pastes, etc.). The P5 strain of the Aspergillus genus was detected at 1,663.49 μg/kg (total AF) and 1,656.22 μg/kg (AFB1) by UPLC, at 1,468.12 μg/kg (total AF) and 1,464.23 μg/kg (AFB1) by HPLC, and at ≥20 μg/kg by ELISA, exhibiting the highest aflatoxin production among the doenjang strains. Similarly, the P7 strain was detected at 1,470.08 μg/kg (total AF) and 1,463.03 μg/kg (AFB1) by UPLC, at 1,056.73 μg/kg (total AF) and 1,056.73 μg/kg (AFB1) by HPLC, and at ≥20 μg/kg by ELISA, demonstrating a high detection level. The MFDS guideline specifies that the aflatoxin level in plant-based materials should be ≤15.0 μg/kg for the sum of B1, B2, G1, and G2 (≤10.0 μg/kg for B1), whereas the CODEX Alimentarius Commission (CAC) that serves as the food standard program of the FAO/WHO specifies that it should be ≤15.0 μg/kg (CODEX, 1995; MFDS, 2022). Therefore, the P5 and P7 strains cannot be used as a fermentation starter owing to the high aflatoxin production, which is above the specified criteria, even though they were isolated from doenjang. Furthermore, the P5 and P7 strains exhibited similar levels of aflatoxin production to the positive controls A. flavus KACC46453, KACC46817, and KACC46449, indicating that they should be reclassified as A. flavus from the previous identification as A. oryzae based on gene sequencing. The P3 and P5 strains with 91.7% homology in the ITS-based phylogenetic tree, as shown in Fig. 2, were differentiated into non-aflatoxigenic and aflatoxigenic strains. Despite the detection of trace amounts or nondetection, the P3 and P4 strains from doenjang and the 40-2 and 83-3 strains from nuruk were demonstrated to produce low levels of aflatoxin compared to the CODEX General Standard for Contaminants and Toxins in Food and Feed. Thus, they were determined to be within the scope of safe food microorganisms for human consumption. In contrast, the commercially used GRAS strains, ATCC1011 and RIB40, were demonstrated to produce trace amounts of aflatoxin at low levels compared to the same CODEX criteria. Because of the lack of criteria for aflatoxin detection in food microorganisms in addition to the criteria for foods, no scientific evidence for the identification of A. flavus based solely on the detection level has been reported thus far. Thus, there is an urgent need for strict criteria to define and distinguish between A. oryzae and A. flavus considering the risk of aflatoxin in the absence of any regulations for identifying fungal strains that produce no aflatoxin as A. orzyae for use among the strains found in fermented foods, including the commercial A. orzyae strain.
Statistical analysis was performed with aflatoxin total contents, and unquantified contents were excluded. Values with the same letter within rows were significantly equal, and different letter within rows were significantly different (p⟨0.05).
Furthermore, AFB1 was detected in all strains by UPLC, whereas B2, G1, and G2 were only partially detected or not detected. In contrast, aflatoxin was not detected in the P4, ATCC1011, RIB40, 40-2, and 83-3 strains by HPLC, despite the low-level detections achieved by UPLC. Moreover, the level of quantification was lower for HPLC than that for UPLC, even at high concentrations. For ELISA, detection was possible in the total aflatoxin range of 2-20 μg/kg. Among all test strains, only the total aflatoxin concentration for P3 and P4 strains were quantified at 1.99 and 2.13 μg/kg, respectively, whereas those of the P5, P7, KACC46453, KACC46817, and KACC46449 strains were quantified at ≥20 μg/kg and those of the ATCC1011, RIB40, 40-2, and 83-3 strains were quantified at ≤1 μg/kg. Moreover, at low aflatoxin concentrations, the high-performance techniques such as UPLC and HPLC produced statistically significant results (p⟨0.05). However, no statistical significance was observed independent of the technique and analytical conditions at concentrations below the LOD (p>0.05).
The findings of this study suggest that the P5 and P7 strains with high levels of aflatoxin detection should be reclassified as A. flavus from the previous identification as A. oryzae, while highlighting the importance of safety evaluation in addition to simple identification for the use of Aspergillus section Flavi strains. Furthermore, among the analytical methods investigated for safety evaluation based on aflatoxin production, UPLC was demonstrated to be superior compared to HPLC for high-accuracy quantification analyses of both high and low concentration samples. In contrast, ELISA, which is an enzyme-based immunoassay, may be effective for rapid analysis of low-level aflatoxin production.
4. Conclusions
This study compared and evaluated the level of aflatoxin detection by UPLC, HPLC, and ELISA with the aim to analyze the aflatoxin production of Aspergillus sp. isolated from doenjang. The test strains isolated from doenjang, nuruk, and other sources were partially sequenced on the ITS and 18S rRNA regions, and the identification results indicated a limitation of accurate differentiation between A. oryzae and A. flavus owing to high sequence homology. Consequently, to identify A. flavus with aflatoxin production, the test strains were pretreated using Immuno-Affinity Column (IAC), and then analyzed by UPLC and HPLC. The methods were validated by estimating the LOD, LOQ, linearity, repeatability, accuracy, and recovery. All three detection methods evaluated in this study produced satisfactory results according to the CODEX or MFDS aflatoxin detection criteria. Notably, the highest accuracy in terms of LOD and LOQ measurements was demonstrated by UPLC. The P5 and P7 strains isolated from doenjang, which were named A. oryzae based on sequencing, exhibited a high level of aflatoxin production at 1,663.49, 1,468.12, and ⟩20 μg/kg and at 1,470.08, 1,056.73, and ⟩20 μg/kg in the quantitative analyses by UPLC, HPLC, and ELISA, respectively, and thus, they were reclassified as A. flavus. However, the P3 and P4 strains (A. oryzae) were demonstrated to produce a trace amount of aflatoxin below the CODEX criterion by all three methods, which were assessed as strains with low aflatoxin production. The findings of this study suggest that any Aspergillus sp. isolated for use as a fermentation starter should be evaluated for aflatoxin production, and that UPLC and HPLC can be used for accurate quantitative analysis, whereas ELISA is an effective enzyme-based immunoassay method for the rapid detection of strains with low-level aflatoxin production that is below the criteria (≤15 ppb).